December 2024 TPJ Editor choice: A night shift for histone methylation in DNA damage control
Highlighted publication:
Diurnal control of H3K27me1 deposition shapes expression of a subset of cell cycle and DNA damage response genes
https://doi.org/10.1111/tpj.17114
A night shift for histone methylation in DNA damage control
Plants fine-tune their physiology to the time of day, largely through dynamic shifts in gene expression. While these shifts are generally attributed to transcription factor activity, an additional layer of regulation comes from chromatin modifications. Covalent histone modifications, collectively referred to as the “histone code”, affect chromatin structure and recruitment of regulatory proteins and thereby determine transcriptional activity.
Histone marks show distinct links to diurnal and circadian rhythms in plants. In the model plant Arabidopsis thaliana, signatures of Histone acetylation and phosphorylation vary between day and night, and many components of the circadian clock are regulated by histone acetylation. In contrast to histone acetylation, which is generally associated with gene activation, histone methylation can either activate or repress gene expression, depending on the site of modification. For example, Histone H3 monomethylation at Lysine residue 27 (H3K27me1) is associated with switched-off genes: it plays a crucial role in constitutive silencing of transposable elements and contributes to the maintenance of heterochromatin and the low expression of some genes within. However, it remained unknown whether H3K27me1 deposition follows diurnal patterns, and how such patterns affect gene function.
In the highlighted publication, Fung-Uceda, Gomez and colleagues observed that H3K27me1 levels fluctuate with the time of day, with higher levels at night than during the day (Fig. 1A), and that this difference was more pronounced under short-day conditions than under long-day conditions. H3K27me1 is deposited by the methyl transferases ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) and ATXR6. In agreement with increased H3K27me1 levels, transcript levels of ATXR5 peaked during the night, while ATXR6 transcript levels remained low throughout the 24-hour period (Fig. 1B).
A full knock-out of both ATXR5 and ATXR6 is lethal, but a hypomorphic atxr5 atxr6 double mutant displays reduced leaf and rosette size. Fung-Uceda et al. found that this phenotype is specific to short-day conditions, aligning with the larger fluctuations in H3K27me1. While H3K27me1 deposition in the atxr5 atxr6 mutant was significantly reduced both at midday and midnight, more genes were differentially expressed at night. Notably, genes with decreased H3K27me1 levels were largely more highly expressed, whereas most genes with lower expression showed no change in H3K27me1 levels and are likely not direct ATXR5/6 targets. These observations support the role of H3K27me1 as a repressive mark, with a more prominent role at nighttime.
Genes more highly expressed in atxr5 atxr6 were enriched for those involved in cell cycle control and DNA damage repair (DDR), and many showed a marked drop in the H3K27me1 signal across their gene body, but not at the transcriptional start site (Fig. 1C). Interestingly, many DDR genes also exhibited rhythmic expression patterns in atxr5 atxr6. Expression of core circadian clock genes remained unchanged in atxr5 atxr6, suggesting that DDR gene activation is directly diurnally gated by H3K27me1 deposition. To test whether the responsiveness to DNA damage varied by time of day, wild-type and atxr5 atxr6 plants were treated with bleomycin, a genotoxic compound that causes DNA double strand breaks. In the wild type, bleomycin-induced DDR gene expression was highest at night, while its pattern and magnitude in atxr5 atxr6 remained unaffected by bleomycin treatment.
Taken together, the findings by Fung-Uceda et al. imply that the response to DNA damage varies with time of day, and that this effect is mediated by H3K27me1. The physiological relevance of this gating mechanism is currently unclear. Fung-Uceda et al. propose that H3K27me1 helps synchronise DDR activity with cellular processes such as DNA replication, during which DDR gene function might be crucial to maintain genome integrity. H3K27me1 might thus function as a “repressive switch” that prevents unnecessary induction of DDR genes at times when the risk of DNA damage is low. Moving forward, corresponding author Crisanto Gutierrez plans to further investigate the link between diurnal control of genome replication and DDR, potentially providing evidence to support the long-standing hypothesis that the cell cycle is diurnally gated to avoid DNA damage during times of hight light exposure.
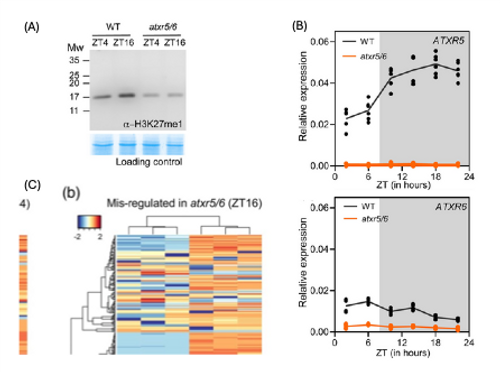
Figure 1. H3K27me1 deposition is diurnally gated.
- Western blot analysis of H3K27me1 levels, detected with anti-H3K27me1 antibody. Coomassie blue staining is shown as a loading control.
- Diurnal time course of ATXR5 and ATXR6 transcript levels in wild-type and atxr5 atxr6 mutant plants grown under short-day conditions.
- Profile of the H3K27me1 signal normalised to total H3 in wild-type and atxr5 atxr6 mutant plants at midday (ZT4) and midnight (ZT16) across genes more highly expressed in atxr5 atxr6.
Figure modified from Fung-Uceda et al.